Allosteric regulation is a phenomenon that the function of the protein changes due to the perturbation at a distant site. Allosteric regulation plays key roles in many biological processes. For example, G protein-coupled receptors, transcription factors and kinases are typical allosteric proteins. Biological systems can use allostery to rapidly up-regulate or down-regulate the activity of target proteins. Therefore, allosteric drugs can be designed to activate or inhibit the desired protein function. Due to their high specificity and low possibility of side effects, allosteric drugs have become one of the major focuses in drug discovery. Though many allosteric site prediction methods have been developed, predicting allosteric regulation type before experimental study remains challenging. The key factors that determine the allosteric regulation type remain elusive.
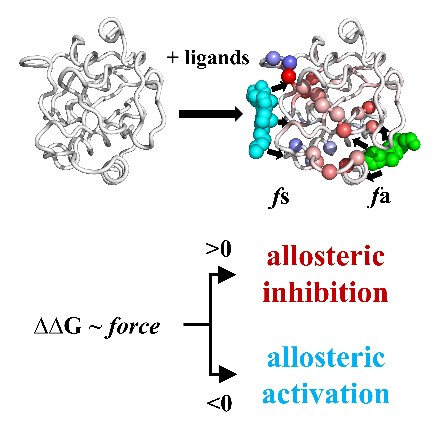
Fig. 1 Ligand-protein interaction controls allosteric regulation type and allosteric pathways
Research groups from the College of Chemistry and Molecular Engineering at Peking University developed an approach that can predict protein allosteric type for the first time. They applied elastic network model and linear response theory to analyze the forces of protein-ligand binding sites and found that allosteric regulation is closely related to the force distributions in ligand binding pockets. They developed the prediction program – AlloType, to predict the direction of allostery and potential allosteric pathways upon allosteric ligand binding for protein function. The prediction success rate of AlloType on the test set is 81%.
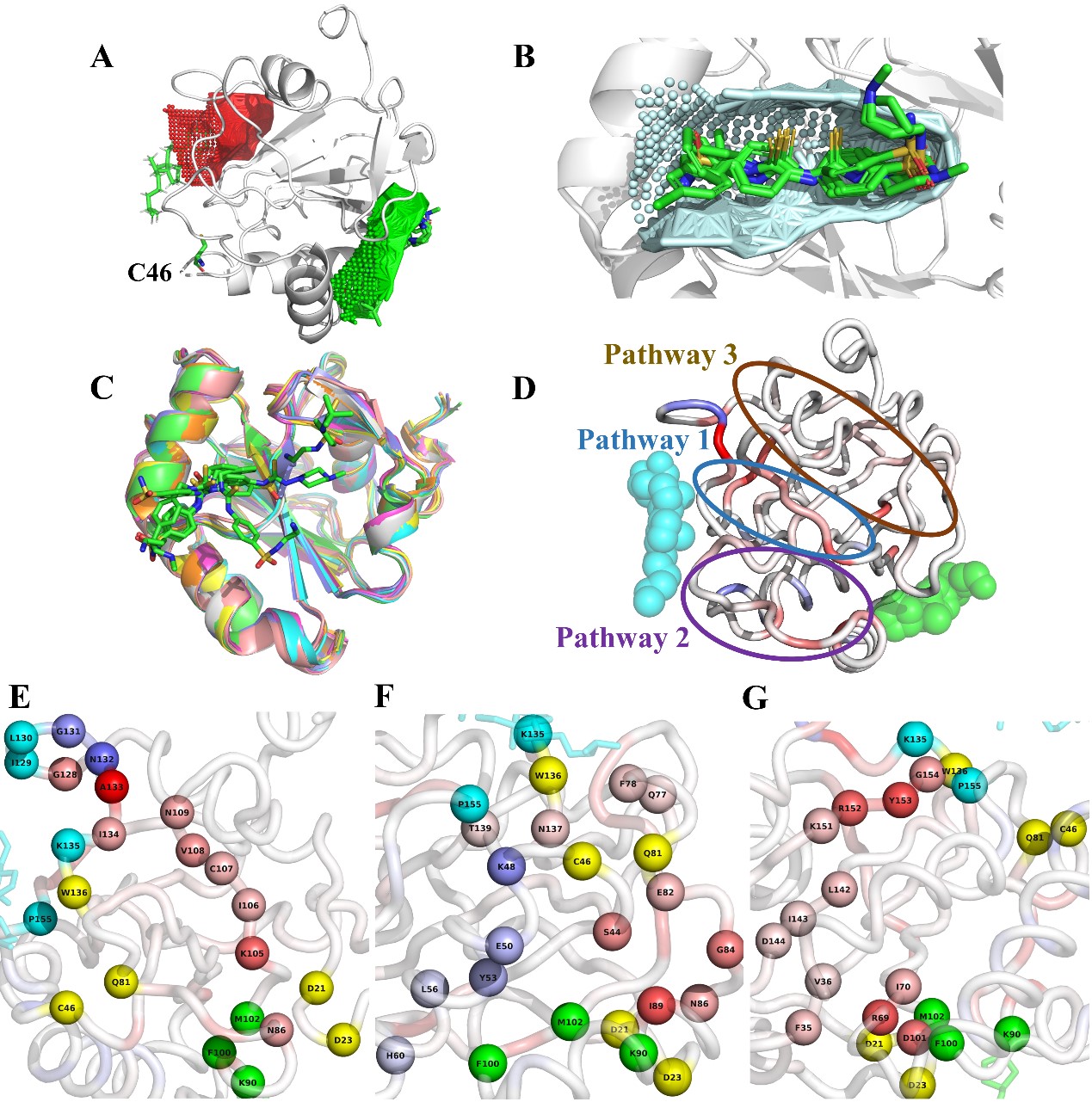
Fig. 2 Prediction of potential allosteric pathways of GPX4 allosteric activator
In early stage of allosteric drug discovery, usually only the free protein structure information is available. To test the application of AlloType in practical allosteric drug discovery programs, they studied GPX4, a key player in ferroptosis. In previous studies, Luhua Lai’s group has developed protein allosteric site prediction methods (J Chem Inf & Mod 2016, 56, 1725; Nucl Acids Res 2018, 46, W374.) and a comprehensive online protein binding site analysis server CavityPlus (http://www.pkumdl.cn/cavityplus), which are widely used in allosteric drug design. They have identified a novel allosteric site and found eight allosteric activators of GPX4 that bind to it (J Med Chem 2019, 62, 266). Based on the modelled complex structures, AlloType can successfully predict the activation effect of these eight molecules on GPX4, and gives three potential allosteric pathways, providing guidance for further molecular optimization.
This work uncovers the relationship between allosteric regulation and the distribution of forces in the ligand binding pocket. And for the first time, the prediction of the allosteric regulation direction of protein function has been realized. AlloType provides a computational tool for the prediction of protein allosteric regulation direction.
The related work was recently published in the Journal of Physical Chemistry Letters (Huang Q, et al. "Allosteric Type and Pathways Are Governed by the Forces of Protein−Ligand Binding", DOI: 10.1021/acs.jpclett.1c01253). The first author is Qiaojing Huang, a PhD student in the College of Chemistry and Molecular Engineering, Peking University, and the corresponding authors are Professor Luhua Lai and Professor Zhirong Liu. The research was funded by the National Natural Science Foundation of China, the Ministry of Science and Technology, the Peking University-Tsinghua Center for Life Sciences, and the Beijing National Laboratory for Molecular Sciences. AlloType can be accessed at https://github.com/qj-Huang/AlloType.
Original link for the paper: https://pubs.acs.org/doi/10.1021/acs.jpclett.1c01253